Now Reading: Detection of Offender DNA Following Skin-to-Skin Contact with a Victim
-
01
Detection of Offender DNA Following Skin-to-Skin Contact with a Victim

Detection of Offender DNA Following Skin-to-Skin Contact with a Victim
Introduction
DNA is one of the best and powerful sources of evidence in the field of forensic science because it contains a large amount of genetic information which can be used for individualization. A few years back, molecular biology and genetics were not advanced and sensitive enough to analyze the traced quantity of DNA but today with the advancement of technology, the methods of analysis are more efficient and sensitive that can even profile a very small quantity of DNA from few DNA containing cells.[1] Now the techniques are so developed that they can more precisely evaluate the origin of DNA. Touch DNA is a traced quantity of DNA obtained in the biological materials transferred from a donor to a person or a thing after physical contact. According to Locard’s Principle of Exchange, every contact leaves a trace. [2] Similarly, when the offender and victim come in contact, the traces of their epithelial cells, sweat, oils are exchanged. The latest technology can determine DNA from seven to eight cells from the outermost layer of skin. [3] The DNA profile of the suspect can be compared with the DNA profile generated from the skin cells obtained from the victim. If it matches, then the presence of the suspect at the crime scene and his contact with the victim is confirmed. This method can be very helpful in the investigation of cases where there is an absence of other important evidence such as biological fluids, etc. Touch DNA is also called epithelial DNA. The amount of Touch DNA found at a crime scene can even be less than 100 picograms. Hence it is also called Trace DNA or Low copy DNA. [3]
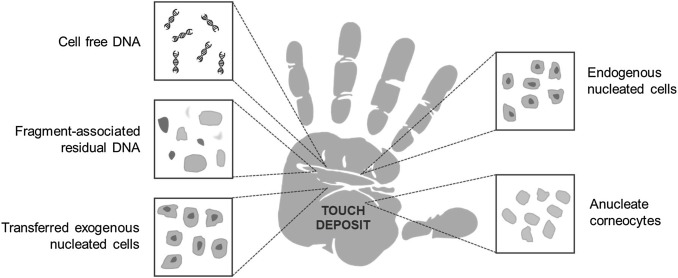
Fig. 1. Concept map of potential sources of DNA deposited by touch/ handling. It is currently well established that individuals may leave behind detectable DNA when they handle items, but the anatomical origin of the DNA remains unsolved. It is possible that the DNA typically recovered from handled items in forensic scenarios comes from nucleated cells from hands, anucleated cells from hands, nucleated cells transferred onto hands from elsewhere, residual cell fragments (including free nuclei) from hands or from outside a cellular architecture in sweat on hands or residual transferred body fluids. [4]
Image source: https://images.app.goo.gl/3uKgwRoEVz2BiSedA
The skin is the largest body organ consisting of epithelial cells, secretions of sweat glands, and oil glands. These cells act as a good potential source of DNA. Skin cells are nucleated, and each human cell contains about 5 picograms of nuclear DNA. [5] PCR can produce a profile of less than 100 picograms of DNA. Therefore 20 cells are more than enough for DNA typing. Touch DNA can be obtained from anything (the victim’s body or any object) that has been touched by the donor. The amount of sloughed epithelial cells deposited on the surface may depend upon their adhering power, amount of surface areas in contact, type of contact, time of contact, and on an individual (some people are good shedders and some are not).
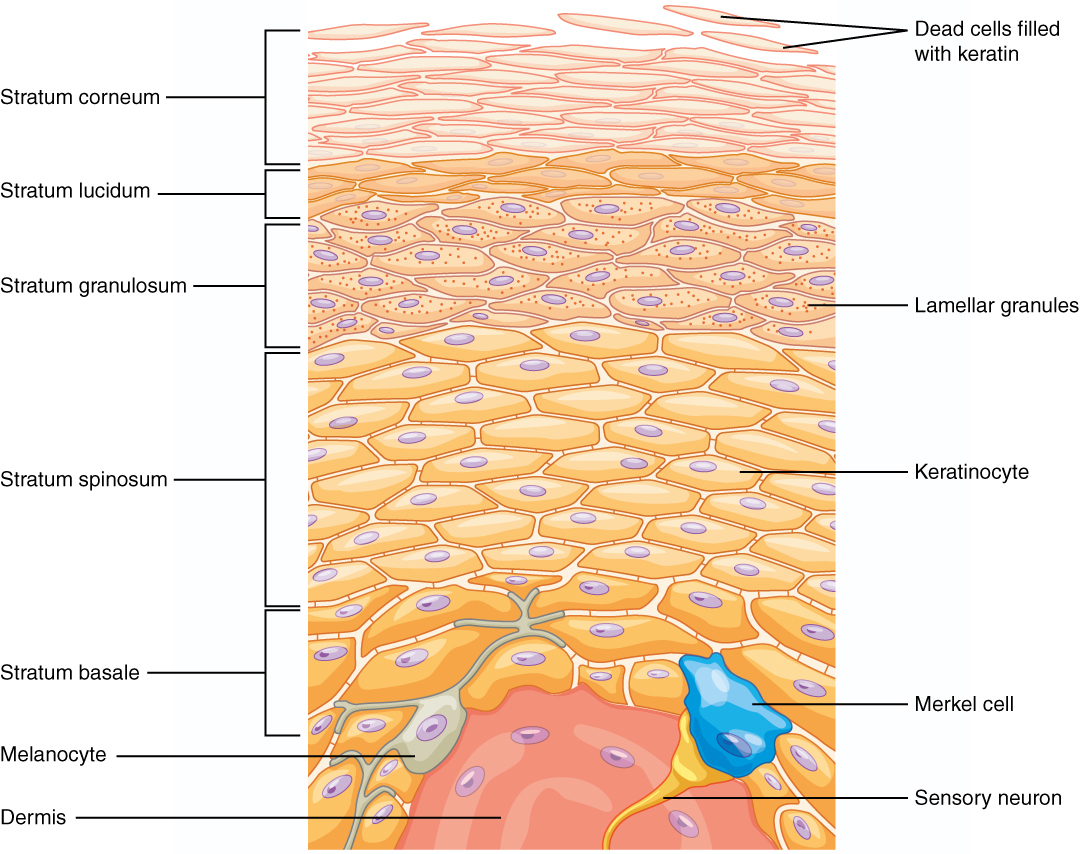
Fig. 2. Epidermal layers in the thick skin of the palms of the hands distinguished by cell types. The process of terminal differentiation occurs as cells move up to the outermost layer of skin, generating an outer layer consisting of flattened keratinocytes lacking nuclei (corneocytes) made mostly of keratin filaments joined tightly together in lipid- and protein-dense, highly cross-linked matrix. [4]
Image source: https://images.app.goo.gl/tw5uFDkVsbxnSEtU8
Good and Bad Shedders
The amount of touch DNA deposited on the victim and his garments may vary from offender to offender. Hence leads to the classification of individuals as good or bad at depositing DNA on handled objects [6] i.e good and bad shedders. The Good shedders are also called good epithelial cell donors (sloughers) and bad shedders are called poor epithelial cell donors (non-sloughers). [5] Good shedder might shed a large amount of different skin cells as mentioned in Fig. 2 whereas bad shedders might shed less or no cells.
DNA Profiling
1. Sampling
Crime scene investigators use various methods of samplings such as wet/dry swabbing, cutting, scraping, and tape lifting. In the swabbing method, a wet sterile cotton swab is rubbed against the surface which is hard and non-porous such as glass, metals, etc. followed by a dry swab to collect possible skin cells. [7] The cutting method is used when the area of interest is a soft item, fabric, or garment. In this method, a piece of the item with the possibility of the presence of cells is cut and packed in a tube. In the scraping method, a sterile scalpel blade is used to scrape the fragments from soft and porous items or surfaces. The tape lifting method focuses on the areas where perpetrators have had maximum contact. An adhesive tape is placed on the surface and pulled off. This method allows sampling of larger surface areas. The sample should be collected from the hands, neck, face, garments, and other areas where there is a possibility of contact. While collecting trace DNA, the sampling area has huge importance. The whole investigation can be meaningless if the sample is not collected from the specific areas of contact. Hence determining the areas of contact can be a tough task. For comparison, the reference sample from the suspect can be obtained from the suspect’s saliva, blood, any other body fluid, or body tissue.
2. DNA Extraction
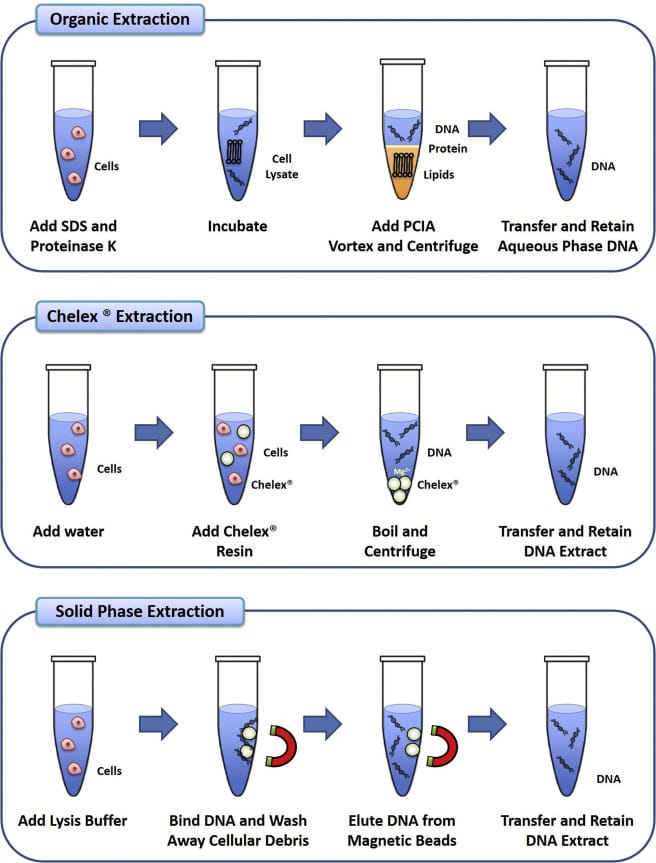
DNA can be extracted from the collected samples (both questioned and reference) using various extraction methods such as organic extraction (phenol-chloroform), chelex extraction, solid-phase extraction (silica gel-based).
- Organic Extraction: also known as phenol-chloroform extraction is a technique in which SDS (sodium dodecyl sulfate) and Proteinase K are added to the sample for the digestion of proteins and cellular components. Then a mixture of phenol: chloroform: isoamyl alcohol (25: 24: 1) is added to break down lipids and to promote the debris into the organic phase, leaving DNA into an aqueous phase. [8] Later the centrifugation followed by ethanol precipitation allows to recover and concentrate DNA from an aqueous phase. This method is highly efficient because it extracts double-stranded DNA but is very time-consuming and uses toxic chemicals.
- Chelex Extraction: Chelex is a chelating agent which has an affinity towards polyvalent metal ions. In this method, Chelex resin is added to the sample solution which causes heat denaturation of cells. High temperature leads to the release of DNA into the solution and chelex resin binds to magnesium ions which stops activation of deoxyribonucleases and prevents the degradation of DNA. This method is cheap and rapid but the presence of debris may result in the degradation of extracted DNA.
- Solid-phase Extraction: This is similar to organic extraction only the phenol is replaced by insoluble particulate materials (silica-coated magnetic beads) which work in a similar manner as that of phenol. Firstly, magnetic beads are added to the sample solution which binds to nucleic acids. Then the solution containing beads is washed to remove the contaminants. Later the beads are washed with chaotropic salts to elute the DNA and free it from the silica-coated magnetic beads. This method works on the principle of selective binding of DNA to the silica beads.
Illustration comparing three major methods of DNA Extraction [8]
3. DNA Quantification
After extracting the DNA, it is very important to know the amount of DNA that has been extracted from the samples. It is done by DNA quantification in which the amount, concentration, and purity of DNA are determined. DNA quantification can be done using various methods such as UV-spectrophotometry, Fluorescence-based dye staining, gel electrophoresis.
- UV-spectrophotometry– The maximum absorbance of nucleic acids (DNA and RNA) is observed at 260nm and of proteins is at 280nm. The ratio of absorbance of nucleic acid to protein is used to measure the purity of DNA. It should be in between 1.8- 2.0, greater than 2 indicated low quality of DNA. This is an easy and quick method.
- Fluorescence-based dye staining– This method uses dyes that bind to ssDNA, dsDNA, or RNA and produce fluorescence after excitation. The amount of DNA is directly proportional to the fluorescent signal.
- Gel electrophoresis– The DNA sample is loaded in the wells of agarose gel and an electric field is applied. The DNA fragments move on the basis of their molecular weight. Smaller fragments move faster as compared to larger ones. Once done with electrophoresis, the gel slab is stained with ethidium bromide and is visualized under UV. It reveals the band intensity of DNA which is proportional to the concentration of DNA. The location of the band indicated the size of the DNA fragment.
4. DNA Amplification
After quantification, the DNA is amplified using PCR (Polymerase Chain Reaction) technique. It is a technique which can make copies of a particular section of DNA. Firstly, the double-stranded DNA is denatured to single stranded under high temperature (94℃). Then at low temperature (50-56℃), DNA primers complementary to the sticky ends of ssDNA are attached. This process is called Annealing. Again at a high temperature (72℃), the Taq polymerase enzyme bring about elongation. This cycle is repeated 20-40 times to obtain the amplified DNA.
5. DNA Profiling
DNA profiling is also called DNA fingerprinting. DNA profiles can be obtained by various methods such as RFLP, RAPD, AFLP, STR, etc.
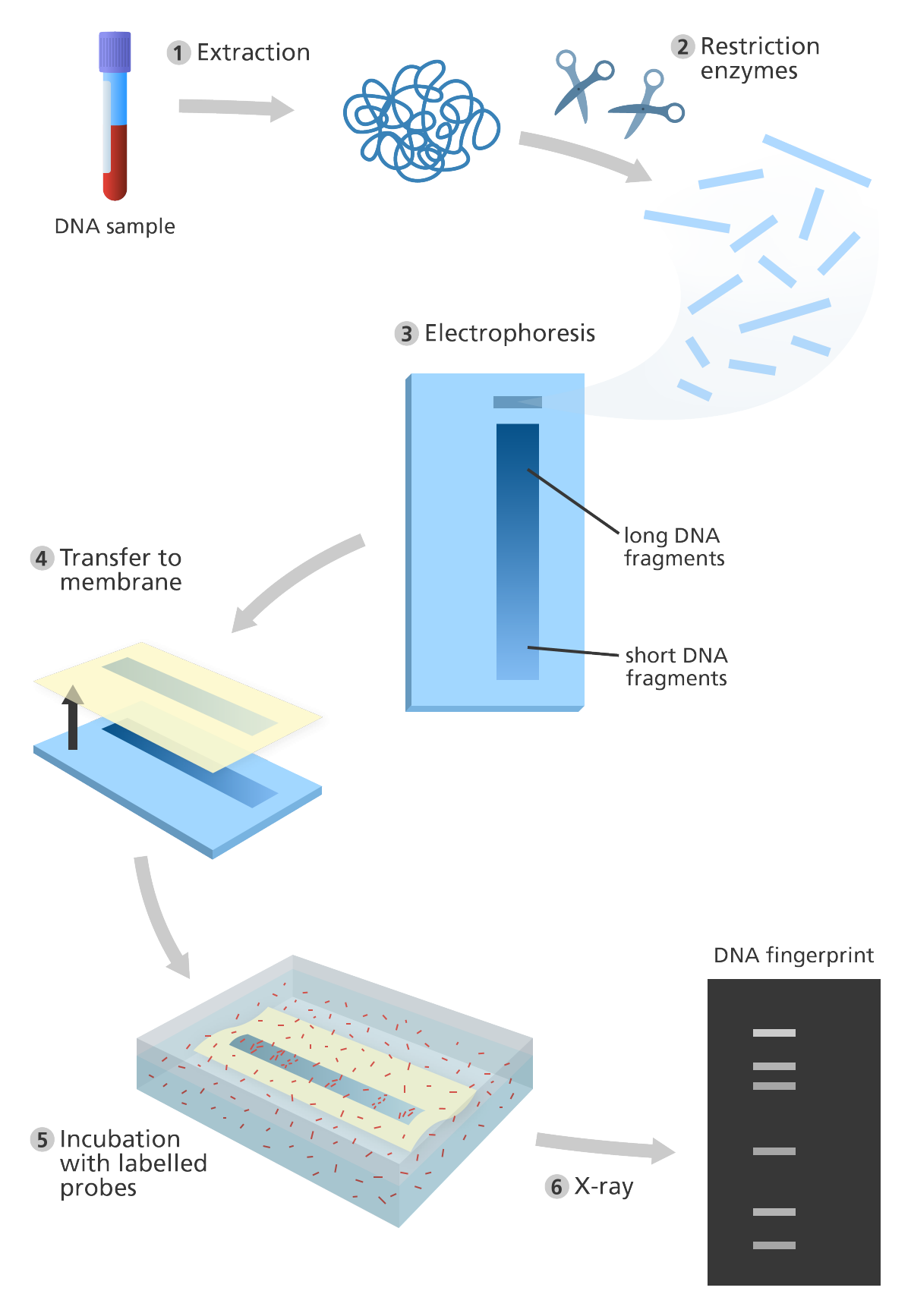
- RFLP (Restriction Fragment Length Polymorphism)- RFLP is found in both coding and non-coding regions of DNA. It consists of various steps such as fragmentation by restriction enzymes, sorting the fragments by gel electrophoresis, neutralizing DNA by soaking the gel in alkaline solution, transferring the DNA fragments to a nylon membrane for southern blotting, hybridization of ssDNA with radioactive labelled complementary DNA probes followed by its exposure to X-Rays to obtain the DNA profile.
- RAPD (Random Amplified Polymorphic DNA)- This method is dependent on the amplification of genomic DNA by primers of an arbitrary nucleotide sequence. RAPD markers are dominant hence makes it difficult to distinguish between homozygous and heterozygous sequences at a particular locus. This method lacks blotting and hybridization. The arbitrary primers are added to the extracted DNA and then amplified using PCR. The fragments are separated by gel electrophoresis and then stained with EtBr (Ethidium Bromide) and visualized under UV light to obtain the DNA profile.
- AFLP (Amplified Fragment Length Polymorphism)- AFLP can detect polymorphism in different genomic regions simultaneously. Firstly, the extracted DNA is digested with restriction enzymes followed by ligation of restriction half site-specific adaptors to the sticky ends of all restriction fragments. Later, the fragments are amplified by PCR primers that have restriction-specific sequences. The next step is an electrophoretic separation of amplicons on a gel matrix and visualization.
- STR (Short Tandem Repeats)- Short tandem repeats (microsatellites) are a group of 2-7 base pairs that are repeated sequences of DNA which repeat numerous times in a head-tail manner and are unique to individuals. The extracted DNA is amplified by primer sequences which bind to STR flanking regions. This step is followed by gel electrophoresis, staining, and scanning using a gel imager. The results can be obtained in the form of an electropherogram or a text-based table. This STR profile can then be compared with the reference sample’s STR profile.
Fig. 3. Illustration showing the steps in DNA fingerprinting [9]
Image source: Genome Research Limited
DNA Profile Match Results
When the band profiles from both the questioned sample and reference sample match completely then the DNA is said to have the same origin i.e it belongs to the same individual. (Fig.4. Suspect 2 shows match)
When no single band from both the questioned and reference sample match, then it is called as a mismatch and it shows that the DNA belongs to two different persons and hence the suspect is excluded. (Fig.4 Suspect 3 shows mismatch)
When few bands from both the question and reference sample match, then it is called as a partial match. It shows that the suspect is not the source of a questioned sample but there is a possibility that the close relative of the suspect might be the source of the questioned sample. (Fig. 4. Suspect 1 shows partial match)
Fig. 4. Illustration of partial match (Suspect 1), match (Suspect 2), and mismatch (Suspect 3).
Image source- National Human Genome Research Institute [10]
Analysis of Skin cells
Along with the DNA profile, the analysis of epithelial cells can reveal important information related to the donor such as medication used by him, the risk for certain diseases, presence of illicit drugs or psychotropic substances. This information can give a brief idea about the offender’s lifestyle, habits, drug use, physical health, and psychological state. Thus it can act as a very useful piece of information in an investigation which might help to narrow down the search of offenders and elimination of innocents.
Conclusion
Detection of offender DNA from the touch DNA obtained on victim’s body or garments after a skin-to-skin contact depends on the type of donor whether he is a good shedder or bad. Other factors like time of contact, type of contact, adhering power of shedding epithelial cells also play an important role in touch DNA analysis. It is impossible to prevent the shedding of seven to eight cells on the crime scene which are not noticeable to the naked eyes. Advancement in the technology of trace DNA analysis can make its gateway to the culprit even through these seven to eight cells. Hence no offender can escape the punishment. This technology is so sensitive that it can even include the cells from the innocent person to the crime scene. Contamination is also a very big problem in generating false identification. Hence a thorough examination of circumstantial facts, other physical and corroborative evidence related to the case should be done and then the conclusion should be drawn.
References
- Helmus, J., Bajanowski, T., Poetsch, M. (2016). DNA transfer- – a never ending story. A study on scenarios involving a second person as carrier. International Journal of Legal Medicine, 130(1); 121-125. https://doi.org/10.1007/s00414-015-1284-1
- Sessa, F., Salerno, M., Bertozzi, G. et.al. (2019). Touch DNA: impact of handling time on touch deposit and evaluation of different recovery techniques: An experimental study. Scientific Reports, 9(1), 9542. https://doi.org/10.1038/S41598-019-46051-9
- Mary, L. (2017). Touch DNA and Chemical Analysis of Skin Trace Evidence: Protecting Privacy While Advancing Investigations, William & Mary Bill of Rights Journal, 26(2). [Last accessed on- 08/05/2021] Available from- https://scholarship.law.wm.edu/wmborj/vol26/iss2/2
- Burrill, J., Daniel, B., Frascione, N. (2018). A Review of Trace “Touch DNA” Deposits: Variability Factors and an Exploration of Cellular Composition. Forensic Science International: Genetics. https://doi.org/10.1016/j.fsigen.2018.11.019
- Wickenheiser, R. A. (2002). Trace DNA: a review, discussion of theory, and application of the transfer of trace quantities of DNA through skin contact. Journal of Forensic Sciences, 47(3); 442-450 https://doi.org/10.1520/JFS15284J
- Phipps, M., Petricevic, S. (2007). The tendency of individuals to transfer DNA to handled items. Forensic Sci. Int. 168(2); 162-168 https://doi.org/10.1016/j.forsciint.2006.07.010
- Williamson, AL. (2012). Touch DNA: Forensic Collection and Application to Investigations. J. Assoc. Crime Scene Reconstr., 18(1); 1-5
- Phenol Chloroform Extraction. Trends in Food Science & Technology, 2021. ScienceDirect [online]. [Last accessed- 29/05/2021] Available- https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/phenol-chloroform-extraction
- “yourgenome,” 2 June 2016. [Online]. [Last accessed on- 08/05/2021]. Available: https://www.yourgenome.org/facts/what-is-a-dna-fingerprint
- “National Human Genome Research Institute,” 28 Jan 2016. [Online]. [Last accessed on 08/05/2021]. Available: https://www.genome.gov/genetics-glossary/DNA-fingerprinting